-Search query
-Search result
Showing 1 - 50 of 54 items for (author: kim & cy)
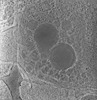
EMDB-28138: 
3D reconstruction of the apical complex of Plasmodium falciparum (3D7) free merozoite
Method: electron tomography / : Segev-Zarko L, Sun SY, Kim CY
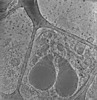
EMDB-28141: 
3D reconstruction of the apical complex of Plasmodium falciparum (3D7) free merozoite
Method: electron tomography / : Segev-Zarko L, Sun SY, Kim CY
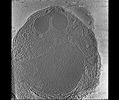
EMDB-28142: 
3D Reconstruction of Plasmodium falciparum (3D7) free merozoite
Method: electron tomography / : Segev-Zarko L, Sun SY, Kim CY

EMDB-28125: 
Plasmodium falciparum merozoites apical 2-ring units
Method: subtomogram averaging / : Sun SY, Pintilie GD

EMDB-28126: 
Toxoplasma apical rings
Method: subtomogram averaging / : Sun SY, Pintilie GD

EMDB-28139: 
Toxoplasma gondii apical complex (ionophore stimulated)
Method: electron tomography / : Segev-Zarko L, Sun SY, Kim CY, Egan ES, Chiu W, Boothroyd JC

EMDB-28140: 
Toxoplasma gondii apical complex (non-stimulated)
Method: electron tomography / : Segev-Zarko L, Sun SY, Chiu W, Boothroyd JC

EMDB-27438: 
Cryo-EM structure of SARS-CoV-2 Beta (B.1.351) spike protein in complex with VH domain F6
Method: single particle / : Zhu X, Saville JW, Mannar D, Berezuk AM, Subramaniam S

EMDB-27439: 
Cryo-EM structure of SARS-CoV-2 Beta (B.1.351) spike protein in complex with VH domain F6 (focused refinement of RBD and VH F6)
Method: single particle / : Zhu X, Saville JW, Mannar D, Berezuk AM, Subramaniam S

PDB-8di5: 
Cryo-EM structure of SARS-CoV-2 Beta (B.1.351) spike protein in complex with VH domain F6 (focused refinement of RBD and VH F6)
Method: single particle / : Zhu X, Saville JW, Mannar D, Berezuk AM, Subramaniam S

EMDB-24863: 
CryoEM map of modular PKS holo-Lsd14 bound to antibody fragment 1B2, stalled at the condensation step, focused refinement of KS-KS'-ACP domains
Method: single particle / : Bagde SR, Kim CY, Fromme JC

EMDB-24862: 
CryoEM structure of modular PKS holo-Lsd14 bound to antibody fragment 1B2, stalled at the condensation step, consensus refinement
Method: single particle / : Bagde SR, Kim CY, Fromme JC

EMDB-24864: 
CryoEM map of modular PKS holo-Lsd14 bound to antibody fragment 1B2, stalled at the condensation step, focused refinement of LDAT domains
Method: single particle / : Bagde SR, Kim CY, Fromme JC

EMDB-24865: 
CryoEM map of modular PKS holo-Lsd14 bound to antibody fragment 1B2, stalled at the condensation step, focused refinement of LD'AT' domains
Method: single particle / : Bagde SR, Kim CY, Fromme JC

EMDB-24866: 
CryoEM map of modular PKS holo-Lsd14 bound to antibody fragment 1B2, stalled at the condensation step, focused refinement of KR domain
Method: single particle / : Bagde SR, Kim CY, Fromme JC

EMDB-24867: 
CryoEM map of modular PKS holo-Lsd14 bound to antibody fragment 1B2, stalled at the condensation step, focused refinement of DD* and 1B2
Method: single particle / : Bagde SR, Kim CY, Fromme JC

EMDB-24869: 
CryoEM map of modular PKS holo-Lsd14 bound to antibody fragment 1B2, consensus refinement
Method: single particle / : Bagde SR, Kim CY, Fromme JC

EMDB-24870: 
CryoEM map of modular PKS holo-Lsd14 bound to antibody fragment 1B2, focused refinement of KSAT dimer
Method: single particle / : Bagde SR, Kim CY, Fromme JC

EMDB-24871: 
CryoEM map of modular PKS holo-Lsd14 bound to antibody fragment 1B2, focused refinement of LDAT domains
Method: single particle / : Bagde SR, Kim CY, Fromme JC

EMDB-24872: 
CryoEM map of modular PKS holo-Lsd14 bound to antibody fragment 1B2, focused refinement of LD'AT' domains
Method: single particle / : Bagde SR, Kim CY, Fromme JC

EMDB-24873: 
CryoEM map of modular PKS holo-Lsd14 bound to antibody fragment 1B2, focused refinement of KR domain
Method: single particle / : Bagde SR, Kim CY, Fromme JC

EMDB-24874: 
CryoEM map of modular PKS holo-Lsd14 bound to antibody fragment 1B2, focused refinement of DD* and 1B2
Method: single particle / : Bagde SR, Kim CY, Fromme JC

EMDB-24880: 
CryoEM map of modular PKS apo-Lsd14 bound to antibody fragment 1B2, consensus refinement
Method: single particle / : Bagde SR, Kim CY, Fromme JC

EMDB-24868: 
CryoEM structure of modular PKS holo-Lsd14 stalled at the condensation step and bound to antibody fragment 1B2, composite structure
Method: single particle / : Bagde SR, Kim CY, Fromme JC

EMDB-24875: 
CryoEM structure of modular PKS holo-Lsd14 bound to antibody fragment 1B2, composite structure
Method: single particle / : Bagde SR, Kim CY, Fromme JC

EMDB-11616: 
Cryo-EM structure of the SARS-CoV-2 spike protein bound to neutralizing sybodies (Sb23)
Method: single particle / : Hallberg BM, Das H

PDB-7a25: 
Cryo-EM structure of the SARS-CoV-2 spike protein bound to neutralizing sybodies (Sb23)
Method: single particle / : Hallberg BM, Das H

EMDB-11617: 
Cryo-EM structure of the SARS-CoV-2 spike protein bound to neutralizing sybodies (Sb23) 2-up conformation
Method: single particle / : Hallberg BM, Das H

PDB-7a29: 
Cryo-EM structure of the SARS-CoV-2 spike protein bound to neutralizing sybodies (Sb23) 2-up conformation
Method: single particle / : Hallberg BM, Das H

EMDB-22829: 
Human Tom70 in complex with SARS CoV2 Orf9b
Method: single particle / : QCRG Structural Biology Consortium

PDB-7kdt: 
Human Tom70 in complex with SARS CoV2 Orf9b
Method: single particle / : QCRG Structural Biology Consortium

EMDB-11002: 
CryoEM structure of bovine cytochrome bc1 in complex with a tetrahydroquinolone inhibitor
Method: single particle / : Muench SP, Johnson R, Amporndanai K, Atonyuk S

EMDB-0554: 
Cryo-EM Structure of the Lysosomal Folliculin Complex (FLCN-FNIP2-RagA-RagC-Ragulator)
Method: single particle / : Fromm SA, Young LN, Hurley JH

EMDB-0556: 
Cryo-EM Map of the active Ragulator-RagA-RagC Complex
Method: single particle / : Yokom AL, Fromm SA, Hurley JH
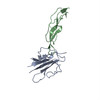
PDB-5ty4: 
MicroED structure of a complex between monomeric TGF-b and its receptor, TbRII, at 2.9 A resolution
Method: electron crystallography / : Weiss SC, de la Cruz MJ, Hattne J, Shi D, Reyes FE, Callero G, Gonen T

PDB-5k7n: 
MicroED structure of tau VQIVYK peptide at 1.1 A resolution
Method: electron crystallography / : de la Cruz MJ, Hattne J, Shi D, Seidler P, Rodriguez J, Reyes FE, Sawaya MR, Cascio D, Eisenberg D, Gonen T

PDB-5k7o: 
MicroED structure of lysozyme at 1.8 A resolution
Method: electron crystallography / : de la Cruz MJ, Hattne J, Shi D, Seidler P, Rodriguez J, Reyes FE, Sawaya MR, Cascio D, Eisenberg D, Gonen T

PDB-5k7p: 
MicroED structure of xylanase at 2.3 A resolution
Method: electron crystallography / : de la Cruz MJ, Hattne J, Shi D, Seidler P, Rodriguez J, Reyes FE, Sawaya MR, Cascio D, Eisenberg D, Gonen T

PDB-5k7q: 
MicroED structure of thaumatin at 2.5 A resolution
Method: electron crystallography / : de la Cruz MJ, Hattne J, Shi D, Seidler P, Rodriguez J, Reyes FE, Sawaya MR, Cascio D, Eisenberg D, Gonen T

PDB-5k7r: 
MicroED structure of trypsin at 1.7 A resolution
Method: electron crystallography / : de la Cruz MJ, Hattne J, Shi D, Seidler P, Rodriguez J, Reyes FE, Sawaya MR, Cascio D, Eisenberg D, Gonen T

PDB-5k7s: 
MicroED structure of proteinase K at 1.6 A resolution
Method: electron crystallography / : de la Cruz MJ, Hattne J, Shi D, Seidler P, Rodriguez J, Reyes FE, Sawaya MR, Cascio D, Eisenberg D, Gonen T

PDB-5k7t: 
MicroED structure of thermolysin at 2.5 A resolution
Method: electron crystallography / : de la Cruz MJ, Hattne J, Shi D, Seidler P, Rodriguez J, Reyes FE, Sawaya MR, Cascio D, Eisenberg D, Gonen T

EMDB-8216: 
MicroED structure of tau VQIVYK peptide at 1.1 A resolution
Method: electron crystallography / : de la Cruz MJ, Hattne J

EMDB-8217: 
MicroED structure of lysozyme at 1.8 A resolution
Method: electron crystallography / : de la Cruz MJ, Hattne J, Shi D, Seidler P, Rodriguez J, Reyes FE, Sawaya MR, Cascio D, Eisenberg D, Gonen T

EMDB-8218: 
MicroED structure of xylanase at 2.3 A resolution
Method: electron crystallography / : de la Cruz MJ, Hattne J

EMDB-8219: 
MicroED structure of thaumatin at 2.5 A resolution
Method: electron crystallography / : de la Cruz MJ, Hattne J, Shi D, Seidler P, Rodriguez J, Reyes FE, Sawaya MR, Cascio D, Eisenberg D, Gonen T

EMDB-8220: 
MicroED structure of trypsin at 1.7 A resolution
Method: electron crystallography / : de la Cruz MJ, Hattne J, Shi D, Seidler P, Rodriguez J, Reyes FE, Sawaya MR, Cascio D, Eisenberg D, Gonen T

EMDB-8221: 
MicroED structure of proteinase K at 1.6 A resolution
Method: electron crystallography / : de la Cruz MJ, Hattne J, Shi D, Seidler P, Rodriguez J, Reyes FE, Sawaya MR, Cascio D, Eisenberg D, Gonen T

EMDB-8222: 
MicroED structure of thermolysin at 2.5 A resolution
Method: electron crystallography / : de la Cruz MJ, Hattne J

EMDB-8472: 
MicroED structure of a complex between monomeric TGF-b and its receptor, TbRII, at 2.9 A resolution
Method: electron crystallography / : Weiss SC, de la Cruz MJ
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
